Browsing ⇒ OBON: Ocean Biomolecular Observing Network by Title
Now showing items 1-20 of 22
-
BBI: an R package for the computation of Benthic Biotic Indices from composition data.
(2018)The monitoring of impacts of anthropic activities in marine environments, such as aquaculture, oil-drilling platforms or deep-sea mining, relies on Benthic Biotic Indices (BBI). Several indices have been formalised to ... -
Best practices for generating and analyzing 16S rRNA amplicon data to track coral microbiome dynamics.
(2023)Over the past two decades, researchers have searched for methods to better understand the relationship between coral hosts and their microbiomes. Data on how coral-associated bacteria are involved in their host’s responses ... -
Collection Standand Operating Procedures: Omics size fractionation. Version 1.00.
(Naples Ecological Research Augmented Observatory (NEREA), Naples, Italy, 2021)The NEREA project aims to promote the development of new indicators for ocean health and new models of ocean microbiomes by coupling traditional physical, chemical, and biological measurements with omics approaches (i.e. ... -
Confocal Microscopy imaging for Opaline Silica Single Cell Skeletons (Polycystines Radiolaria).
(Station Biologique de Roscoff, Roscoff, France, 2018) -
DNA Metabarcoding Methods for the Study of Marine Benthic Meiofauna: A Review.
(2021)Meiofaunal animals, roughly between 0.045 and 1 mm in size, are ubiquitous and ecologically important inhabitants of benthic marine ecosystems. Their high species richness and rapid response to environmental change make ... -
Earth Microbiome Project (EMP) high throughput (HTP) DNA extraction protocol. Version 07112018.
(University of California, San Diego, San Diego, CA, 2018)This protocol accompanies the following publication: Marotz, Clarisse, et al. "DNA extraction for streamlined metagenomics of diverse environmental samples." BioTechniques 62.6 (2017):290-293 -
EMP Sample Submission Guide.
(NOAA Fisheries Southwest Fisheries Research Center, La Jolla,CA, 2018)This protocol was designed for collaborators with the Earth Microbiome Project to contribute samples in a standardized fashion. Raw, frozen samples are submitted in individually labeled tubes, 10 aliquots (identical ... -
Environmental DNA protocol development guide for biomonitoring.
(National eDNA Reference Centre, Canberra, Australia, 2022)The Environmental DNA protocol development guide for biomonitoring (EP guide for biomonitoring) provides harmonised quality control and minimum standard operating procedures. This document is complemented by the Environmental ... -
Environmental DNA test validation guidelines.
(National eDNA Reference Centre, Canberra, Australia, 2022)These Environmental DNA test validation guidelines provide harmonised quality control and minimum standard considerations for developing or validating eDNA/eRNA assays for the purpose of single-species or multi-species ... -
Expanding "Tara" Oceans Protocols for Underway, Ecosystemic Sampling of the Ocean-Atmosphere Interface During Tara Pacific Expedition (2016–2018).
(2019)Interactions between the ocean and the atmosphere occur at the air-sea interface through the transfer of momentum, heat, gases and particulate matter, and through the impact of the upper-ocean biology on the composition ... -
Framing Cutting-Edge Integrative Deep-Sea Biodiversity Monitoring via Environmental DNA and Optoacoustic Augmented Infrastructures.
(2022)Deep-sea ecosystems are reservoirs of biodiversity that are largely unexplored, but their exploration and biodiscovery are becoming a reality thanks to biotechnological advances (e.g., omics technologies) and their ... -
Interoperable vocabulary for marine microbial flow cytometry.
(2022)The recent development of biological sensors has extended marine plankton studies from conducting laboratory bench work to in vivo and real-time observations. Flow cytometry (FCM) has shed new light on marine microorganisms ... -
Marine microbiomes: Towards standard methods and best practices.
(Frontiers Media SA, Lausanne, Switzerland, 2023)The microbiome is key to understanding and sustaining the services that ocean ecosystems provide (Bolhuis et al., 2020). The marine microbiome—an ensemble of microscopic organisms that inhabit water columns, sediments, ... -
Microscope video recording.
(Station Biologique de Roscoff, Roscoff, France, 2019)Protocol to record videos on SBR Microscope. -
Omics community protocols. EuroSea Deliverable D3.19.
(EuroSea Project, , 2023)The aim of the WP3 “Network Integration and Improvements” is to coordinate and enhance key aspects of integration of European observing technology (and related data flows) for its use in the context of international ocean ... -
A practical guide to DNA-based methods for biodiversity assessment.
(Pensoft Advanced Books, Sofia, Bulgaria, 2021)This book represents a synthesis of knowledge and best practice in the field of DNA-based biomonitoring at the time of writing. It has been written with end-users of molecular tools in mind, as well as those who are new ... -
Quanti-iT™ Pico Green dsDNA Assay (Invitrogen P7589).
(Matthew Sullivan Lab, University of Arizona/Ohio State University, Tucson, AZ, 2019)This protocol accompanies the following publication: Ul-Hasan S, Bowers RM, Figueroa-Montiel A, Licea-Navarro AF, Beman JM, Woyke T, et al. (2019) Community ecology across bacteria, archaea and microbial eukaryotes in the ... -
Scanning Electron Microscopy imaging for Opaline Silica Single Cell Skeletons (Polycystines Radiolaria). Version 3.
(Station Biologique de Roscoff, Roscoff, France, 2018)Protocol adapted from: Biard, T., Pillet, L., Decelle, J., Poirier, C., Suzuki, N. and Not, F. (2015) Towards an Integrative Morpho-molecular Classification of the Collodaria (Polycystinea, Radiolaria). Protist 166, ... -
Simple 3D imaging of biogenic silica structures by fluorescence microscopy. Version 2.
(Station Biologique de Roscoff, Roscoff, France, 2018)This protocol describes how to prepare, stain and mount biogenic silica particles (opaline from diatoms, Radiolaria, sponges spicules, phytolith...) for 3D imaging with fluorescence microscopy. The protocol is fast, ... -
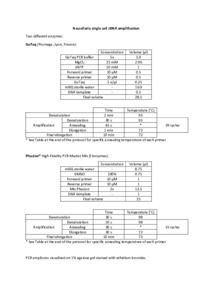
Single cell rDNA amplification of Nassellaria (Radiolaria).
(Station Biologique de Roscoff, Roscoff, France, 2018)
 Repository of community practices in Ocean Research, Applications and Data/Information Management
Repository of community practices in Ocean Research, Applications and Data/Information Management